Andrew D. Marques, PhD, has been passionate about science for as long as he can remember. Formally, he began working on a Biotechnology Certificate Program in 2012. By 2019 he graduated Summa Cum Laude from the University of Florida
with a degree in microbiology and minor in bioinformatics.
In 2024 he completed doctoral studies at the University of Pennsylvania. Andrew has published 5 first/co-first author papers in the past 4 years studying SARS-CoV-2 genome evolution using advanced statistical methods, and AAV capsid engineering using machine learning. Simultaneously, he has enthusiastically engaged in volunteer and paid teaching opportunities for over 8 years. Below are just a few papers he has published.

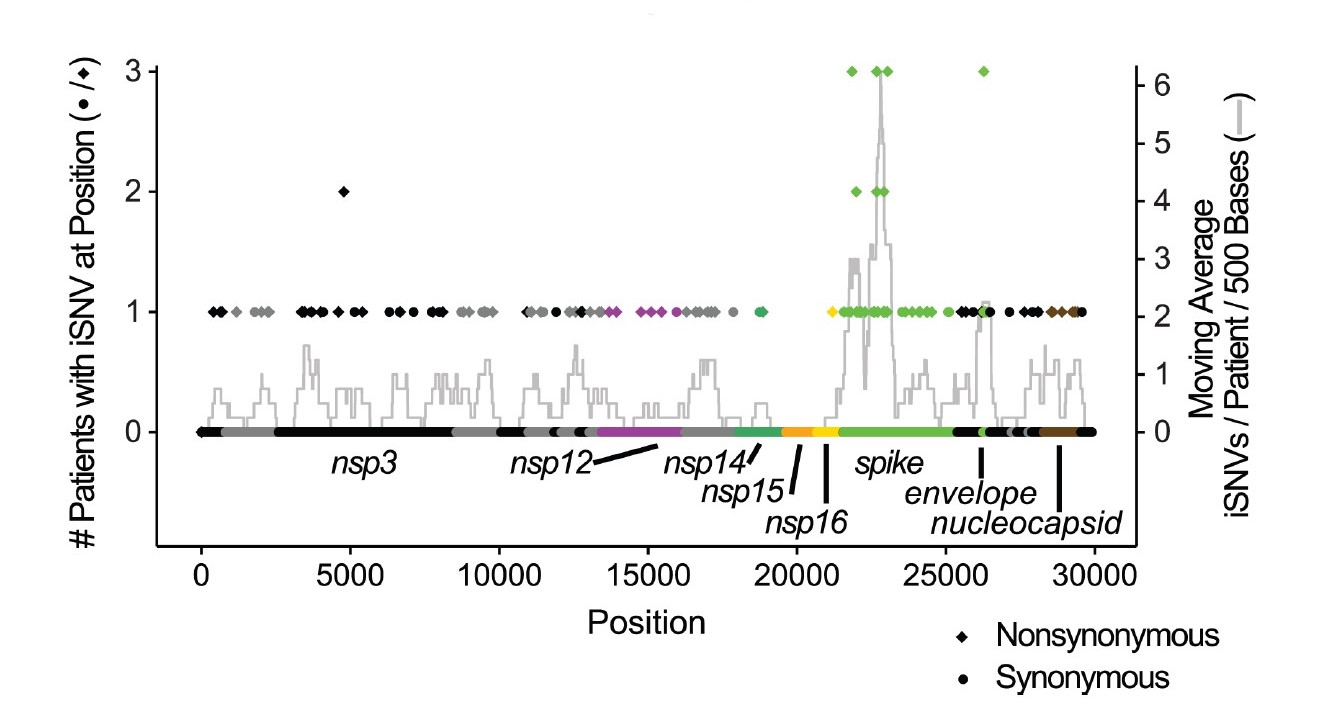
This article, authored by Andrew and colleagues, highlights the within-host evolution of the SARS-CoV-2 virus in immunocompromised patients experiencing long-term infections.
It reveals cases of accelerated evolution rate, especially in the spike coding region. Two substitutions
known to confer anitbody resistance emerged and rose to dominance, highlighting the role of such cases in potentially generating new viral variants.
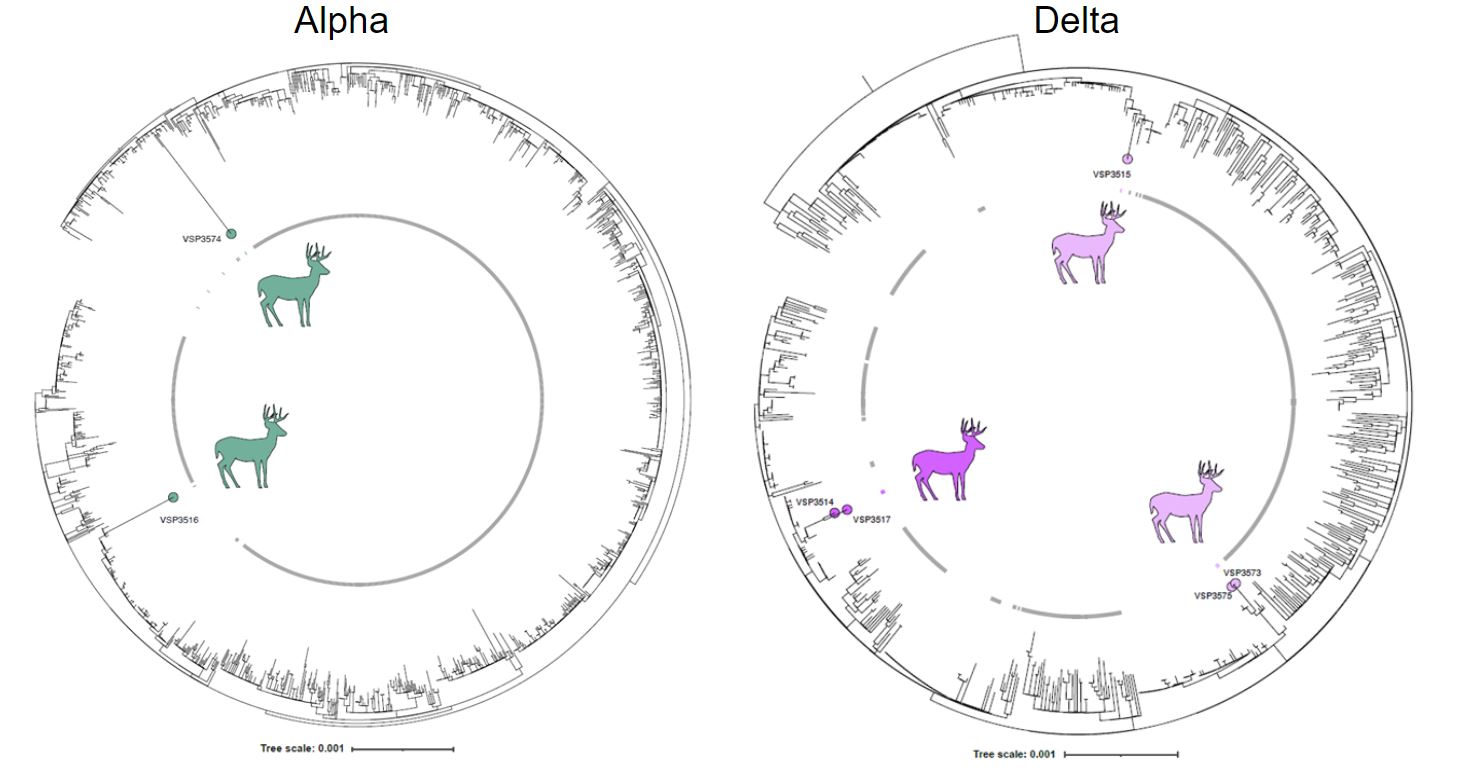
In another first-authored paper, Andrew and collaborators identified multiple new jumps of SARS-CoV-2 from humans to deer,
with alpha lineage persisting in deer after its displacement by delta in humans, and deer-derived alpha variants diverging significantly
from those in humans, suggesting a distinctive evolutionary trajectory in deer. The results provide important information for identifying a
potential new animal reservoir and its management to prevent future infections.
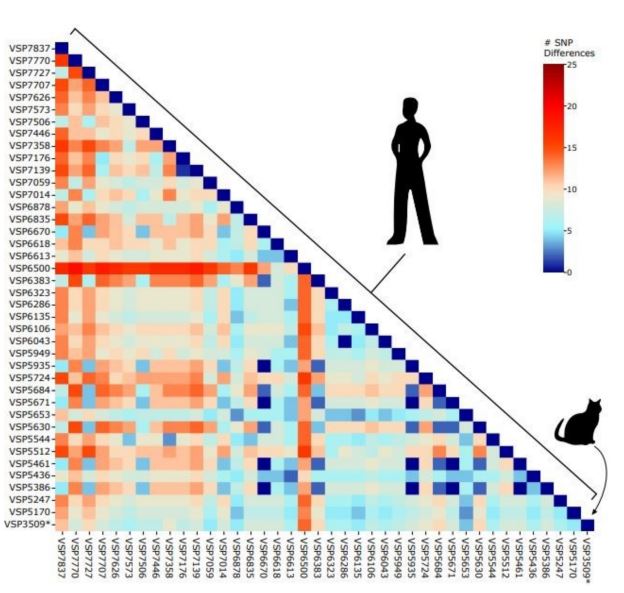
Oliva Lenz and Andrew co-authored the first publication of a delta variant (AY.3) infection of a domestic house cat.
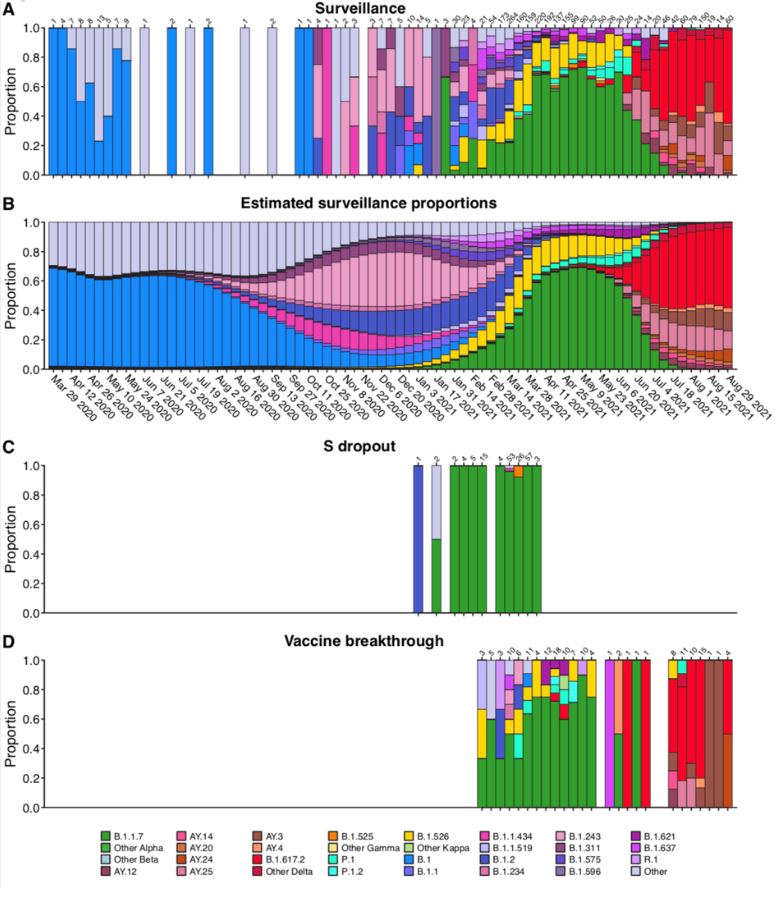
In this first-authored paper, Andrew et al. collected, sequenced, and analyzed viral variants from surveillance of the Delaware Valley and vaccine breakthrough cases,
finding that the delta B.1.617.2 lineage was enriched in breakthrough cases, along with a notable spike substitution, N501Y. With the direction of Scott Sherrill-Mix, Scott and Andrew employed a Bayesian
autoregressive moving average logistic multinomial regression to evaluate vaccine breakthroughs.
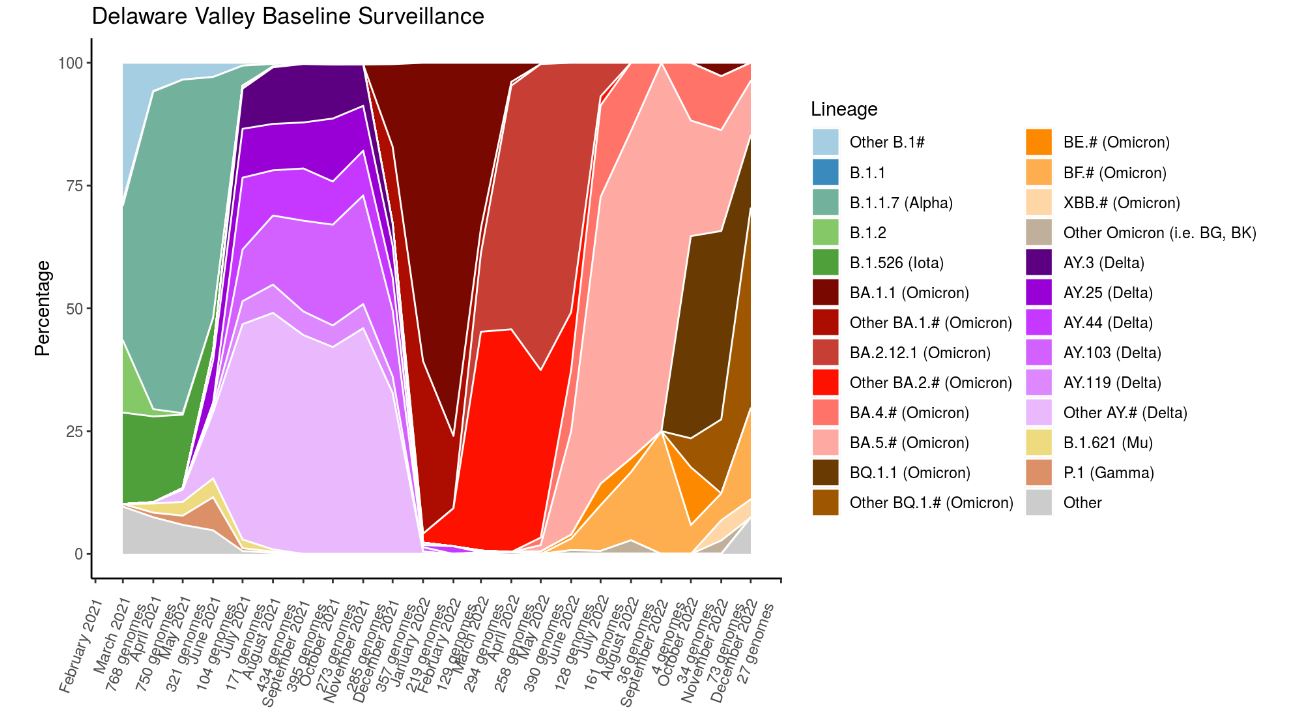
During the height of the pandemic, the Bushman Lab was reporting approximately 10% of all Pennsylvania SARS-CoV-2 surveillance sequences to GISAID and GenBank. Initially built by John Everett,
Andrew began maintaining and updating the SARS-CoV-2 dashboard to report his and the team's efforts to provide real time tracking of viral genomic changes in our region locally.

In this book, Andrew contributes one chapter intended to outline methods and applications for machine learning (ML) in the context of protein engineering. The content of this chapter
is geared toward the biologist’s perspective of ML with an emphasis on experimental design for optimized data collection.
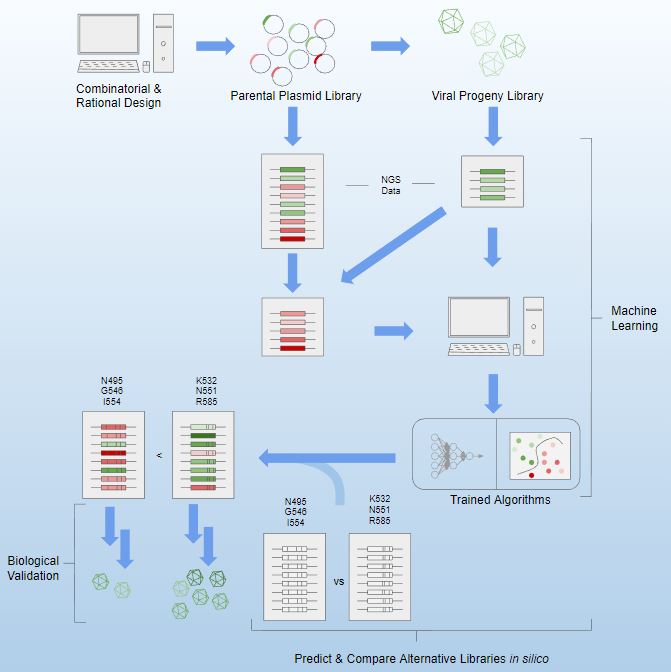
In this first-authored paper, Andrew demonstrates the use of machine learning to analyze next-generation sequencing data of complex capsid libraries for viral gene therapy.
Artificial neural networks and support vector machines were trained to predict whether unknown capsid variants may assemble into viable virus-like structures, with simulated
mutation patterns suggesting the importance of specific amino acids in capsid construction. Comparative libraries were generated using machine learning-derived data to validate
findings and demonstrate predictive power in vector design.
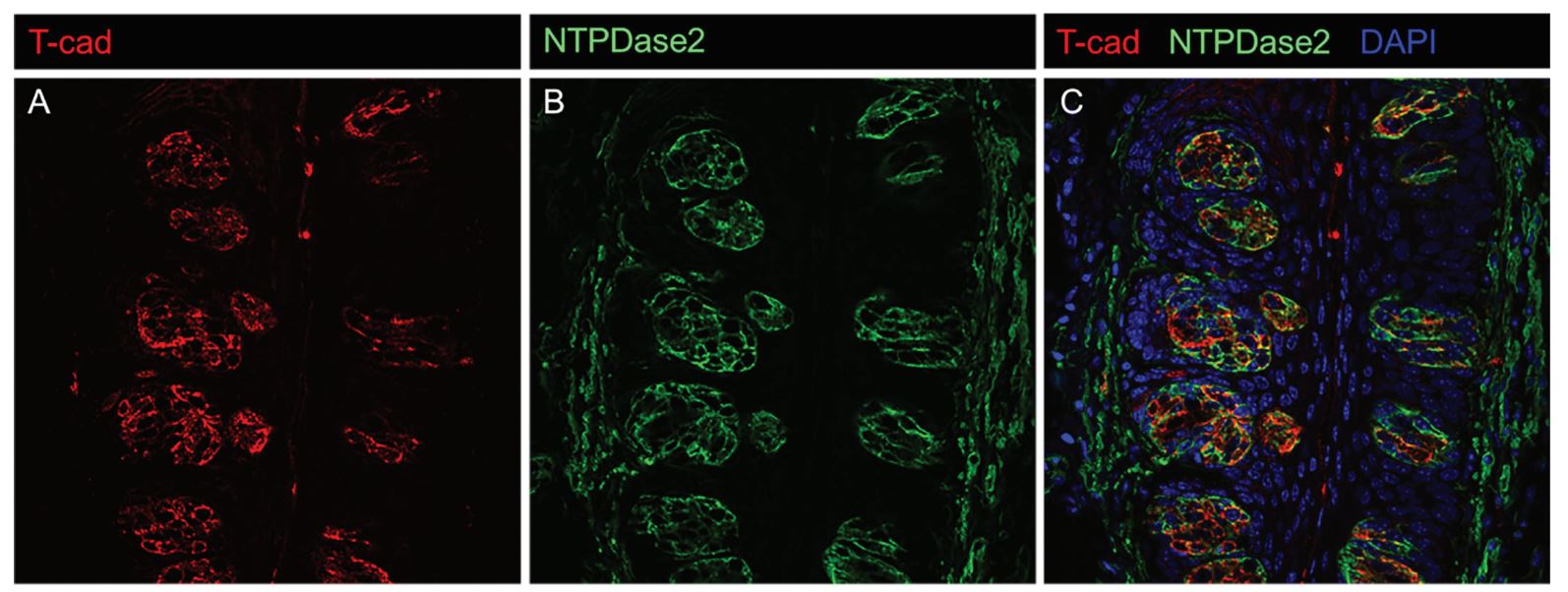
While at the University of Florida, Andrew received training under the direction of Sean Crosson in Sergei Zolotukhin's lab. Andrew is middle author in this work.
This study found that murine taste receptor cells express specific adiponectin receptors and may be targeted by salivary adiponectin, as supported by the presence of adiponectin receptors
in transcriptomic data and immunohistochemical analysis. However, behavioral lick testing did not show a significant difference in mice without adiponectin compared to wild-type mice,
and the impact of salivary adiponectin on taste responsiveness remains unclear.








